6993ef8bcf62f1e9c33a1a7b25b835e6406b0725,Orange/tests/test_svm.py,SVMTest,test_NuSVR,#SVMTest#,50
Before Change
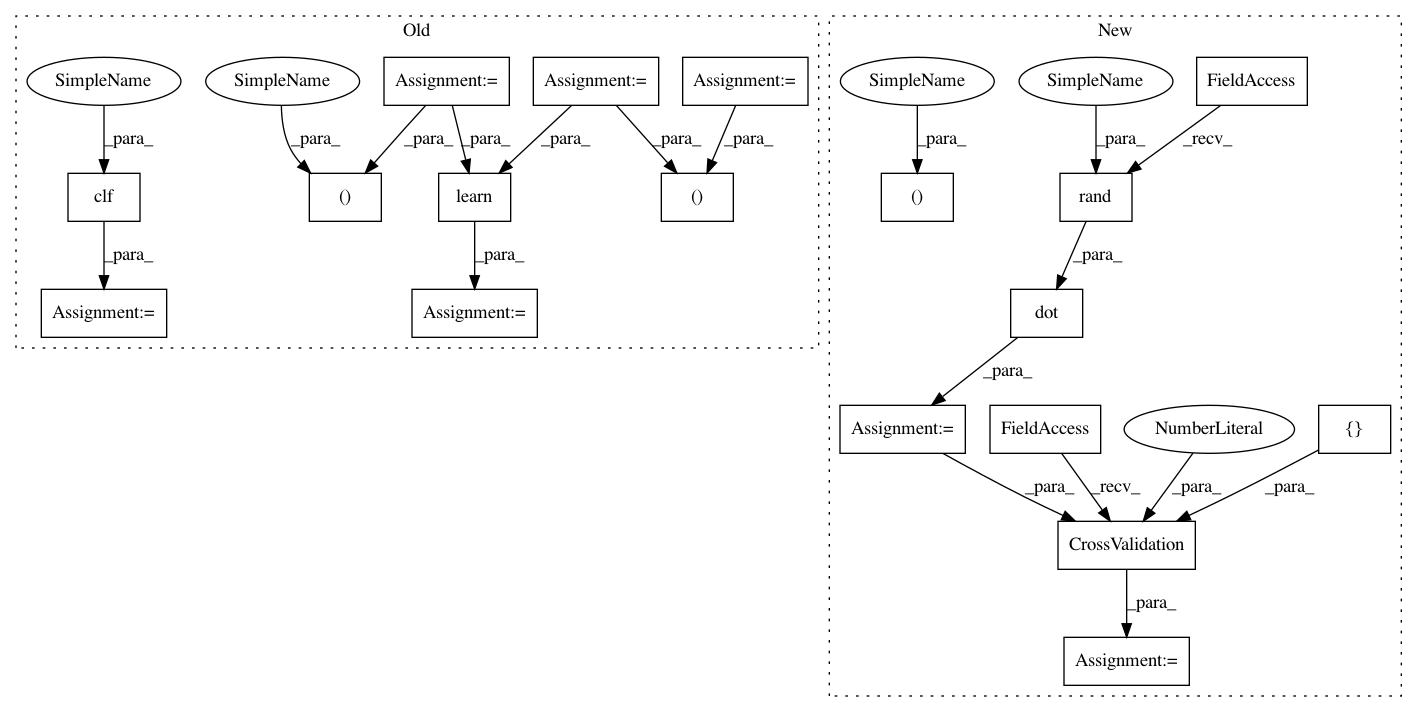
ncols = 5
x = np.sort(10*np.random.rand(nrows, ncols))
y = np.sum(np.sin(x), axis=1).reshape(nrows, 1)
x1 , x2 = np.split(x, 2)
y1 , y2 = np.split(y, 2)
t = Orange.data.Table(x1, y1)
learn = NuSVRLearner(kernel="rbf", C=1e3, gamma=0.1)
clf = learn(t)
z = clf(x2)
self.assertTrue((abs(z.reshape(-1, 1) - y2) < 4.0).all())
def test_OneClassSVM(self):
nrows = 100After Change
self.assertLess(Orange.evaluation.RMSE(res)[0], 0.1)
def test_NuSVR(self):
nrows , ncols = 200, 5
X = np.random.rand(nrows, ncols)
y = X.dot(np.random.rand(ncols))
data = Orange.data.Table(X, y)
learn = NuSVRLearner(kernel="rbf", gamma=0.1)
res = Orange.evaluation.CrossValidation(data, [learn], k=2)
self.assertLess(Orange.evaluation.RMSE(res)[0], 0.1)
def test_OneClassSVM(self):
// TODO: improve the test - what does it check?In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 18
Instances Project Name: biolab/orange3
Commit Name: 6993ef8bcf62f1e9c33a1a7b25b835e6406b0725
Time: 2015-02-16
Author: lan.zagar@fri.uni-lj.si
File Name: Orange/tests/test_svm.py
Class Name: SVMTest
Method Name: test_NuSVR
Project Name: biolab/orange3
Commit Name: bdaae33d6e345738f9d55c777e04ad121f63b1ff
Time: 2015-02-09
Author: lan.zagar@fri.uni-lj.si
File Name: Orange/tests/test_sgd.py
Class Name: SGDRegressionTest
Method Name: test_SGDRegression
Project Name: biolab/orange3
Commit Name: 6993ef8bcf62f1e9c33a1a7b25b835e6406b0725
Time: 2015-02-16
Author: lan.zagar@fri.uni-lj.si
File Name: Orange/tests/test_svm.py
Class Name: SVMTest
Method Name: test_SVR
Project Name: biolab/orange3
Commit Name: 6993ef8bcf62f1e9c33a1a7b25b835e6406b0725
Time: 2015-02-16
Author: lan.zagar@fri.uni-lj.si
File Name: Orange/tests/test_svm.py
Class Name: SVMTest
Method Name: test_NuSVR