cdc6a9b4491b48d9136992c4604792f0057c8bc2,scanpy/tests/plotting.py,,test_stacked_violin,#,103
Before Change
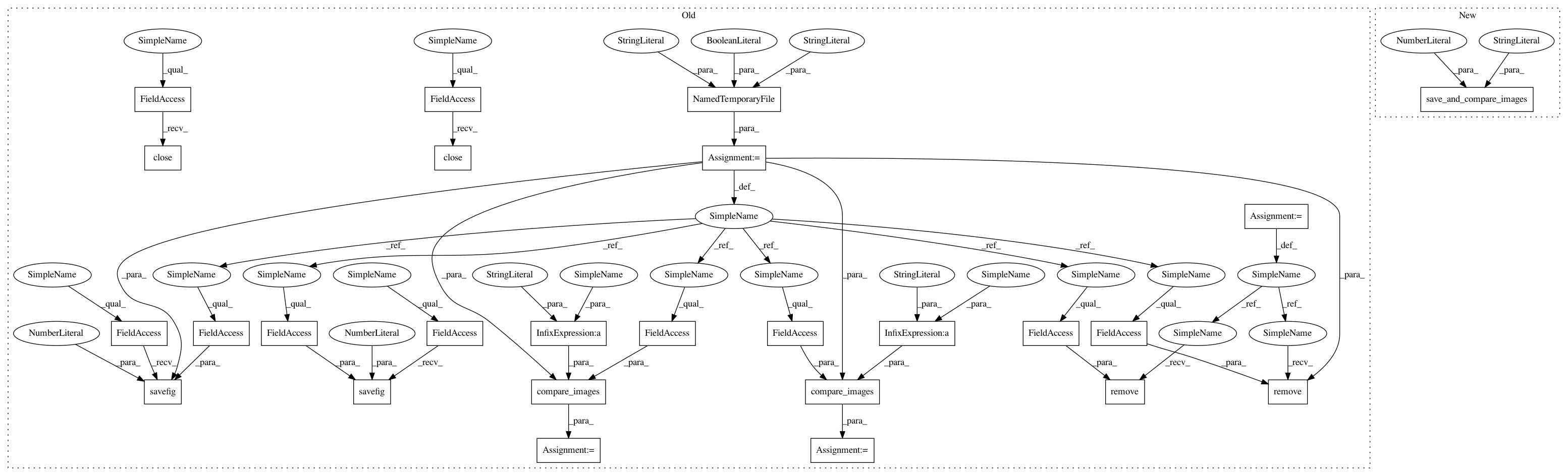
def test_stacked_violin():
adata = sc.datasets.krumsiek11()
outfile = NamedTemporaryFile(suffix=".png", prefix="scanpy_test_stacked_violin_", delete=False)
sc.pl.stacked_violin(adata, adata.var_names, "cell_type", use_raw=False, color="blue", show=False)
pl.title("image may have cut labels.\nThis is ok for test")
pl.savefig(outfile.name, dpi=80)
pl.close()
res = compare_images(ROOT + "/master_stacked_violin.png", outfile.name, tolerance * 2)
assert res is None, res
os.remove(outfile.name)
// test swapped axes
sc.pl.stacked_violin(adata, adata.var_names, "cell_type", use_raw=False,
swap_axes=True, figsize=(3, 5), show=False)
pl.savefig(outfile.name, dpi=80)
pl.close()
re s = compare_images(ROOT + "/master_stacked_violin_swapped_axes.png", outfile.name, tolerance * 2)
assert res is None, res
os.remove(outfile.name)
def test_violin():
sc.pl.set_rcParams_defaults()After Change
sc.pl.stacked_violin(adata, adata.var_names, "cell_type", use_raw=False, color="blue", show=False)
pl.title("image may have cut labels.\nThis is ok for test")
save_and_compare_images("master_stacked_violin", tolerance=15)
// test swapped axes
sc.pl.stacked_violin(adata, adata.var_names, "cell_type", use_raw=False,
swap_axes=True, figsize=(3, 5), show=False)In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 26
Instances Project Name: theislab/scanpy
Commit Name: cdc6a9b4491b48d9136992c4604792f0057c8bc2
Time: 2018-10-09
Author: fidel.ramirez@gmail.com
File Name: scanpy/tests/plotting.py
Class Name:
Method Name: test_stacked_violin
Project Name: theislab/scanpy
Commit Name: cdc6a9b4491b48d9136992c4604792f0057c8bc2
Time: 2018-10-09
Author: fidel.ramirez@gmail.com
File Name: scanpy/tests/plotting.py
Class Name:
Method Name: test_dotplot
Project Name: theislab/scanpy
Commit Name: cdc6a9b4491b48d9136992c4604792f0057c8bc2
Time: 2018-10-09
Author: fidel.ramirez@gmail.com
File Name: scanpy/tests/plotting.py
Class Name:
Method Name: test_matrixplot